
Ordering Information
A Practical Manual of Experimental Biochemistry and Biotechnology (Series II)-GREEN
Price: 500/- PKR
Edition: 2023-2024 (2nd edition)
ISBN: 978-627-7502-05-8 (Print)
Published by Dr. Muhammad Ali
Eighty-eight (88) full-length experiments are included
© Copyright 2023. All rights reserved by Dr. Muhammad Ali
Contact us for purchase and online ordering:
This edition is available on Amazon, Draz, and several other international stores.
Video lectures of almost all the experiments will be uploaded sequentially
Contents of Series II, GREEN
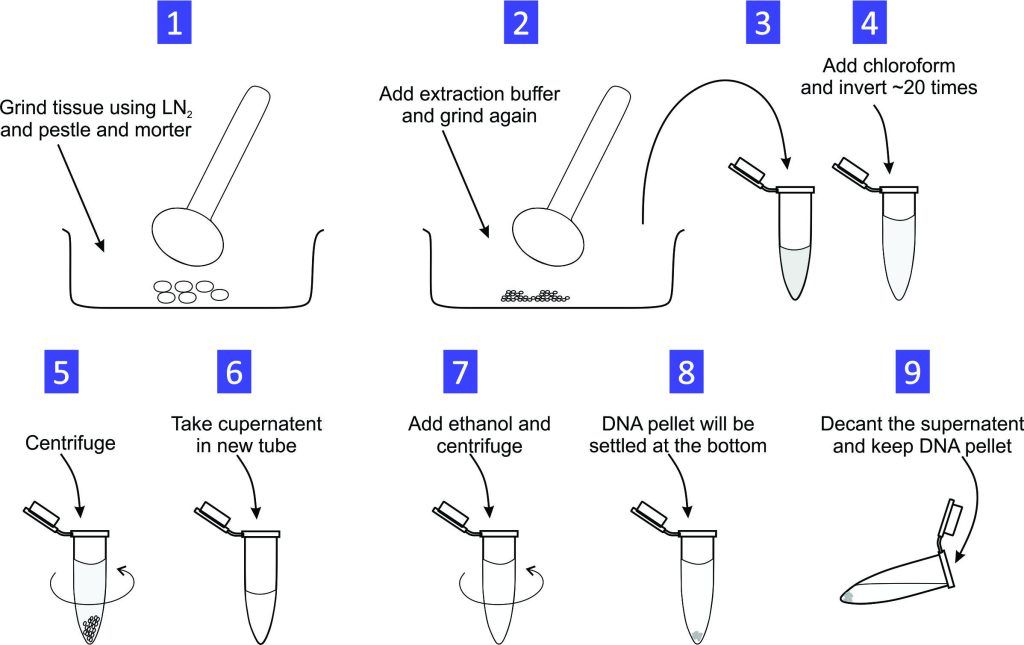
Experiment 1.1- Total Genomic DNA extraction from plant by the CTAB method. 5
Experiment 1.2- DNA extraction from fungi 6
Experiment 1.3- Miniprep extraction of plasmid from bacteria. 6
Experiment 1.4- Total genomic DNA extraction from mouse tail/tissue. 7
Experiment 1.5- Quick tail DNA extraction (Jackson’s lab) 8
Experiment 1.6- DNA extraction from blood. 9
Experiment 1.7- Quantification of DNA and RNA by nanodrop. 9
Experiment 1.8- Designing of primers for endpoint PCR from coding sequence of EI24. 10
Experiment 1.9- Designing of primers for qPCR to check transcription of EI24. 15
Experiment 1.10- Polymerase chain reaction (PCR) for genotyping. 20
Experiment 1.11- Agarose gel electrophoresis of PCR product ~150 bp long. 21
Experiment 1.12- Restricting fragment length polymorphism (RFLP) 22
Experiment 1.13- Polyacrylamide gel electrophoresis (PAGE) of PCR product. 22
Experiment 1.14- Gel extraction protocol (Qiagen Gel Extraction Kit) 24
Experiment 1.15- Purification of PCR product and enzyme-mediated digestion. 24
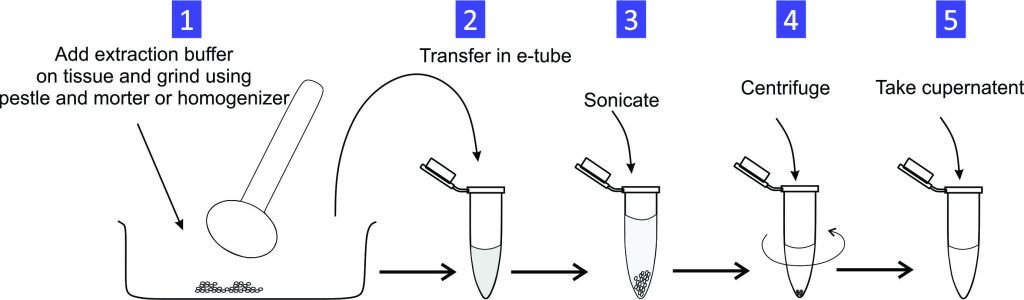
Experiment 1.16- RNA extraction from cultured cells/tissue. 25
Experiment 1.17- Quality test of RNA by agarose gel electrophoresis. 25
Experiment 1.18- Reverse transcriptase polymerase chain (RT-PCR) reaction. 26
Experiment 1.19- Quantitative PCR (qPCR) and operation of the software. 26
Experiment 1.20- Tissue sample lysis for Western blot analysis. 28
Experiment 1.21- Lysis of cultured cells for Western blot analysis. 29
Experiment 1.22- Ammonium sulfate-facilitated protein concentration. 30
Experiment 1.23- Protein quantification by BCA method and protein normalization. 31
Experiment 1.24- SDS-PAGE gel for protein samples. 32
Experiment 1.25- Continuous gradient SDS-PAGE for protein samples. 33
Experiment 1.26- Coomassie Brilliant Blue (CBB) staining for SDS-PAGE. 34
Experiment 1.27- Silver staining for SDS-PAGE gel 34
Experiment 1.28- Western blotting of protein samples obtained from cells and tissues. 35
Experiment 1.29- Protein immunoprecipitation to find protein-protein interaction. 37
Experiment 2.1- Use NCBI to retrieve sequence of PGC1 alfa gene, mRNA, and protein. 40
Experiment 2.2- Translate DNA sequence into RNA sequence. 45
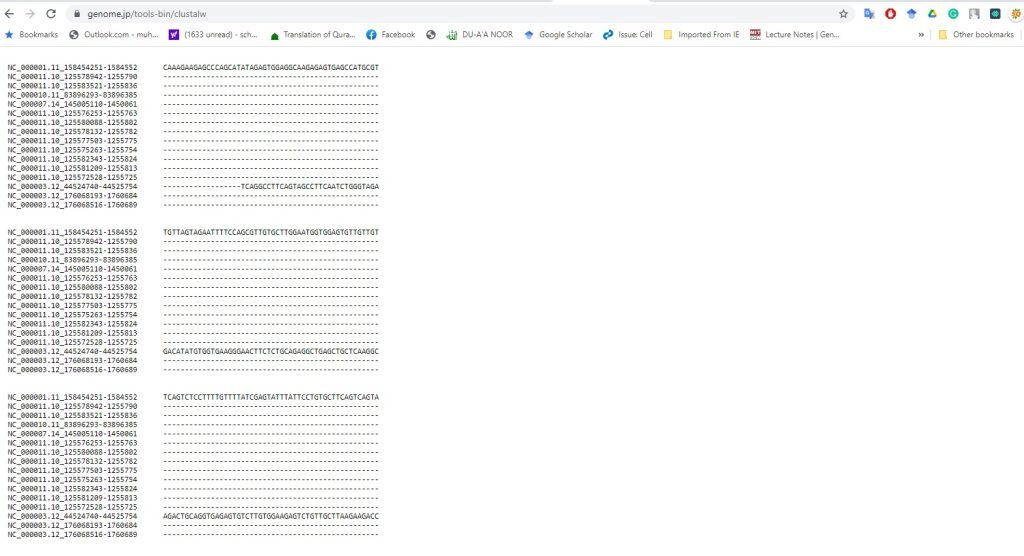
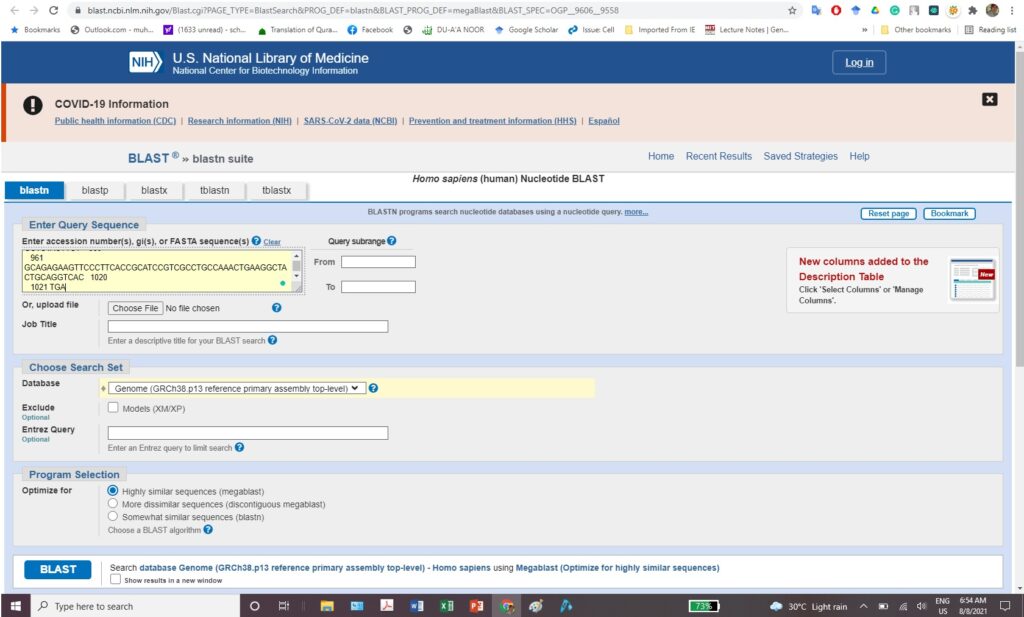
Experiment 2.3- Alignment of two sequences by NCBI 46
Experiment 2.5- Determining of the hydrophobicity or hydrophilicity of protein. 53
Experiment 2.6- Prediction of trans-membrane helix. 55
Experiment 2.7- Secondary structure prediction (alfa-helix and beta-sheets) 56
Experiment 2.8- Prediction of 3D structure of proteins. 57
Experiment 2.9- Codon optimization for expression in bacteria (E. coli) 60
Experiment 2.10- How will you retrieve data and draw phylogenetic tree. 62
Experiment 2.12- Authentication of primers using online tools (oligo analyzer). 73
Experiment 2.13- Prediction of Drug-Receptor interaction Using MOE 2015. 76
Experiment 3.1- Study of cheek cells using methylene blue staining. 81
Experiment 3.2- Study of onion cells and using iodine staining. 81
Experiment 3.3- Observation of fungi cells by lactophenol cotton blue staining. 82
Experiment 3.4- Differentiation between gram +ve and gram –ve bacteria. 82
Experiment 3.5- Stamen hair cells of Tradescantia (light microscopy) 84
Experiment 3.6- Studying different stages of mitosis in onion root tip cells. 84
Experiment 3.7- Study of different stages of meiosis in Tradescantia ohiensis. 85
Experiment 3.8- Different stages of meiosis in the testes of grasshopper. 85
Experiment 3.9- Observing chromosome obtained from blood cells. 86
Experiment 3.10- Observing chromosome obtained from adherent cells (HeLa) 86
Experiment 3.11- Immunohistochemistry (IHC) using anti BrdU antibody. 87
Experiment 3.12- Histology cell shape of skeletal, cardiac, and smooth muscles. 89
Experiment 3.13- White adipose tissue. 90
Experiment 3.14- Liver anatomy and histology. 90
Experiment 3.15- Difference of seed and stem of monocots and dicots. 91
Experiment 3.16- Subculturing adherent mammalian cells. 92
Experiment 3.17- Reviving mammalian cell lines. 93
Experiment 3.18- Cell counting for mammalian cell lines. 93
Experiment 3.19- Cell freezing protocol from ATCC. 94
Experiment 3.20- Thawing of suspension cells. 95
Experiment 3.21- Transfection using lipofectamine. 95
Experiment 3.22- Co-transfection of plasmid DNA and siRNA.. 96
Experiment 3.23- Electroporation of U2OS cell line. 96
Experiment 3.24- Lentivirus production to knockdown TERT mRNA.. 97
Experiment 3.25- Lentivirus Infection and knockdown of TERT mRNA.. 98
Experiment 3.26- Stable cell line expressing EI24 (Transfection method) 99
Experiment 3.27- Preparation of Mouse Embryonic Fibroblasts (MEFs) 100
Experiment 3.28- Luciferase reporter assay. 101
Experiment 3.29- Immunocytochemistry (ICC) for the detection of two proteins. 101
Experiment 3.30- Fluorescently Activated Cell Sorting (FACS) analysis. 102
Experiment 3.31- Determination of cell cycle by FACS. 104
Experiment 3.32- MTT assay for cell viability. 105
Experiment 3.33- Colonogenic assay (crystal violet assay) 105
Experiment 4.1- Preparation of competent cell 108
Experiment 4.2- Transformation of plasmid containing mutated sequence of TRAF6. 109
Experiment 4.3- TA cloning and selection by blue & white colonies. 110
Experiment 4.4- PCR-facilitated introduction of restriction sites and digestion. 111
Experiment 4.5- Restriction of MMTV- ERT2CreERT2 and gel elution. 112
Experiment 4.6- Ligation of the insert into the vector. 113
Experiment 4.7- PCR-facilitated deletion of RING domain from human TRAF6. 114
Experiment 4.8- PCR-facilitated generation of the point mutation (Pro to Leu) 116
Experiment 4.9- PCR-facilitated addition of HA tag at N-term of TRAF6. 117
Experiment 4.10- Production of recombinant insulin. 120
Experiment 4.11- CRISPR/Cas9 for the generation of Tert knockout cell lines and mice. 122
Experiment 4.12- CRISPR/Cas9 to generate Tert knock-in mice at Rosa26 locus. 125
Experiment 4.13- Analyzing mutations generated in Tert knockout cell line/ mice. 127
4.2 Useful numbers for cell culture. 129
4.3 Some restriction enzymes. 130
4.4 Reagents for Biochemistry and Biotechnology lab. 131
4.5- Composition for Tris-glycine SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE) 132
